Importing Suitability
The following instructions describe how to import vegetation suitability values from old pyBRAT runs. Using the script we have provided, this is a quick way to reuse vegetation suitability values that have already been determined.
Step 1
Install Jeff Kunce's dbfpy python package. This allows python to read and comprehend .dbf files. Download and install from this website, or use pip install dbfpy in the command line. This project was written for dbfpy 2.3.1; compatibility with other versions is likely, but not guaranteed.
Step 2
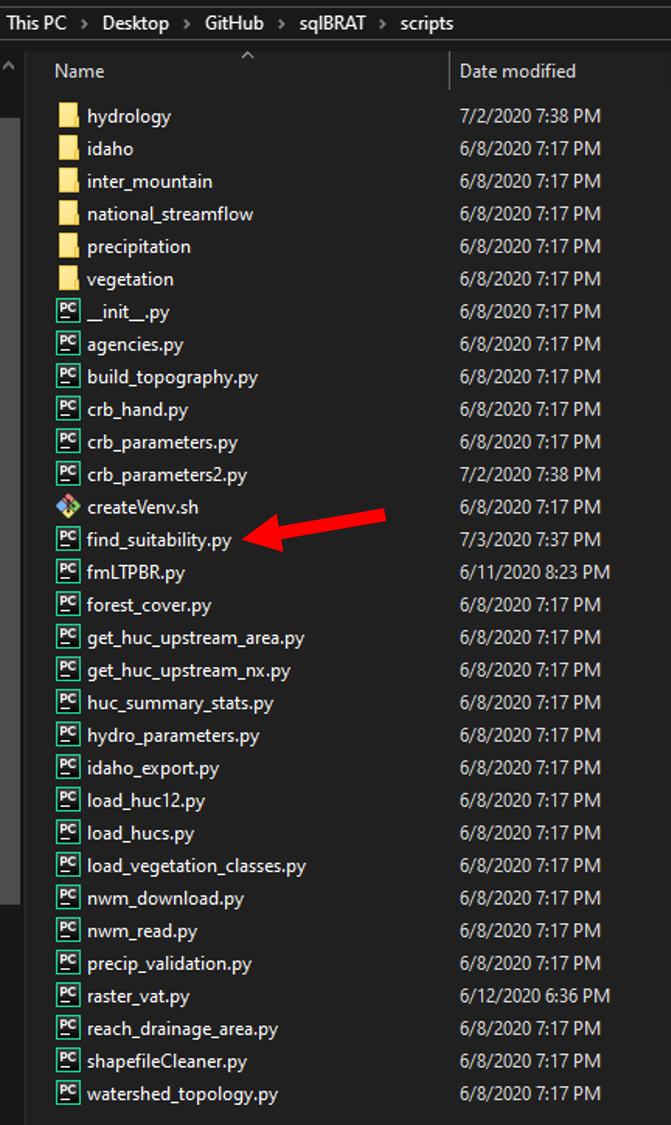
Navigate to the find_suitability.py script. This can be found under sqlBRAT/scripts. Open this in the Python IDE of your choice. For this example we'll be using PyCharm.
Step 3
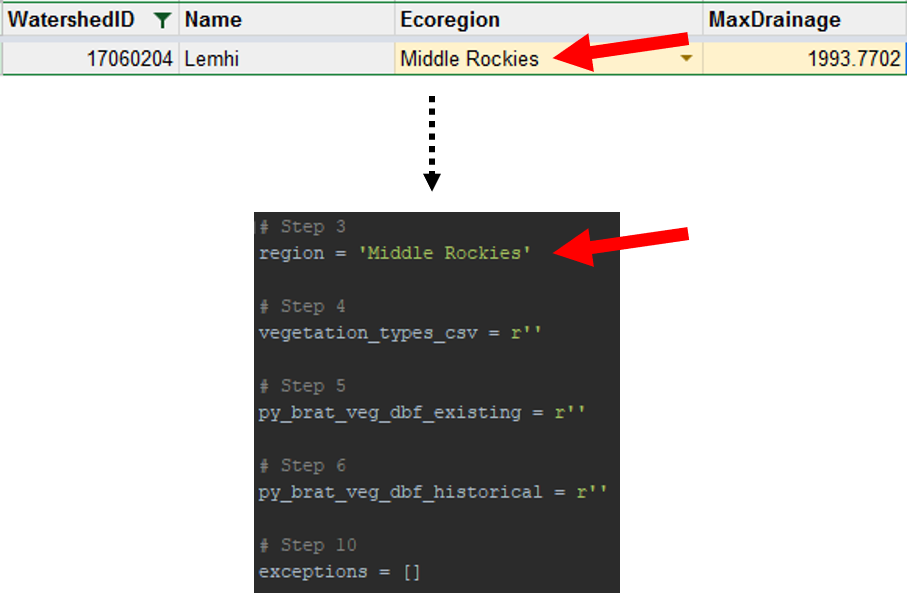
Find your watershed's region within the BRAT Parameters Google Sheet. To do this, navigate to the Watersheds tab. Then, filter the worksheet to the eight digit HUC that you are calibrating. This can be done by clicking the button to the right of the column heading *WatershedID* in the top, leftmost cell. Type your HUC code and click enter to filter the rows. Look at the *Ecoregion* column to determine your watershed's region. Populate the script's first parameter (region) with the path to that file.
Step 4
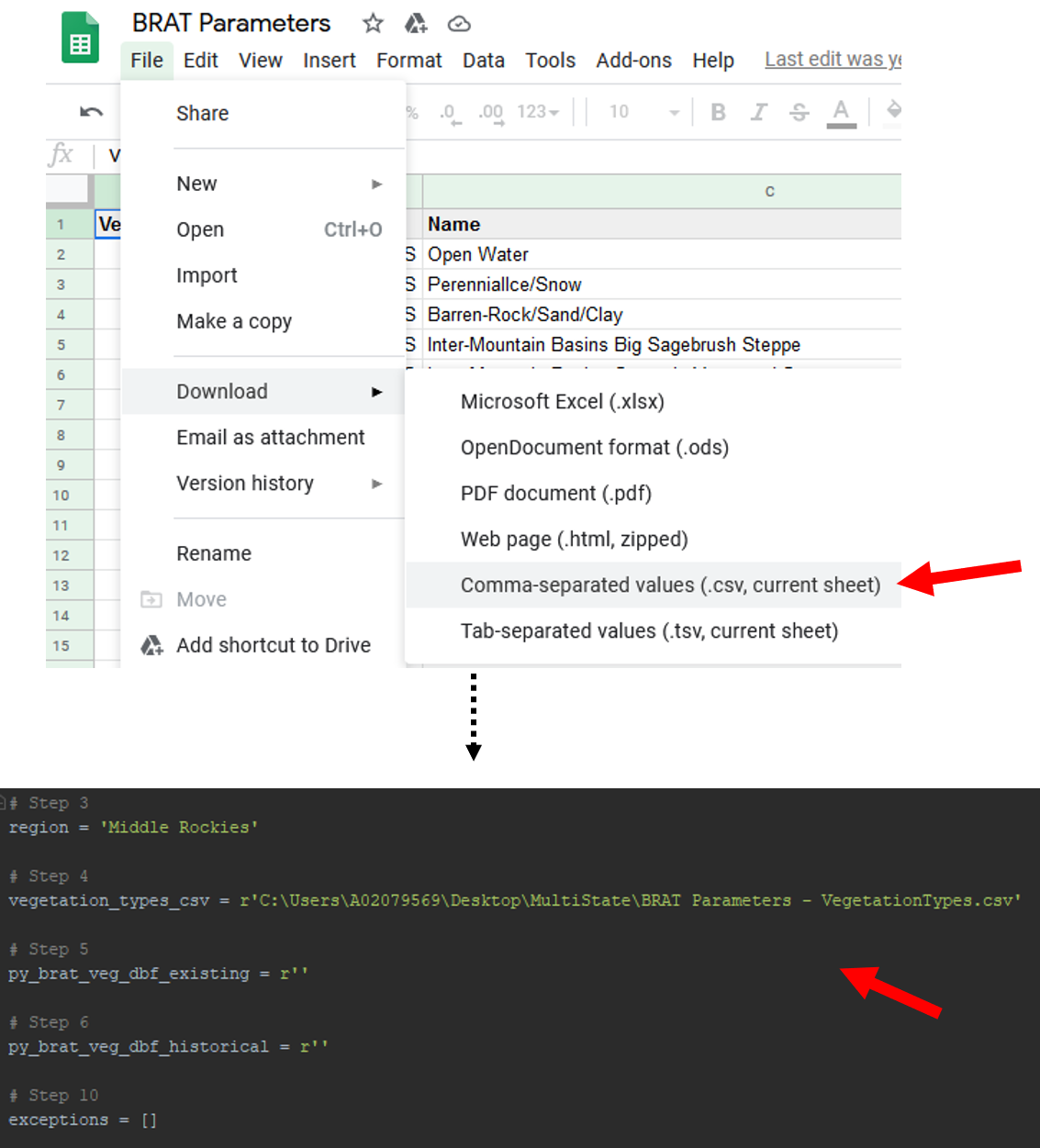
Download the most recent version of the VegetationTypes sheet within the BRAT Parameters Google Sheet. To do this, navigate to the VegetationTypes tab. Then, go to File -> Download -> Comma-separated values (.csv, current sheet). Once the csv downloads, place it somewhere that is easy to navigate to. Populate the script's second parameter (vegetation_types_csv) with the path to that file.
Step 5
Open up your old pyBRAT project to find the existing vegetation raster's .dbf file. This is typically found in Inputs -> 01_Vegetation -> 01_ExistingVegetation -> Ex_Veg_01. Find the Find the file that ends in .dbf. This will typically be something like LANDFIRE_200EVT.vat.dbf or LANDFIRE_140EVT.vat.dbf. Populate the script's third parameter (py_brat_veg_dbf_existing) with the path to that file.
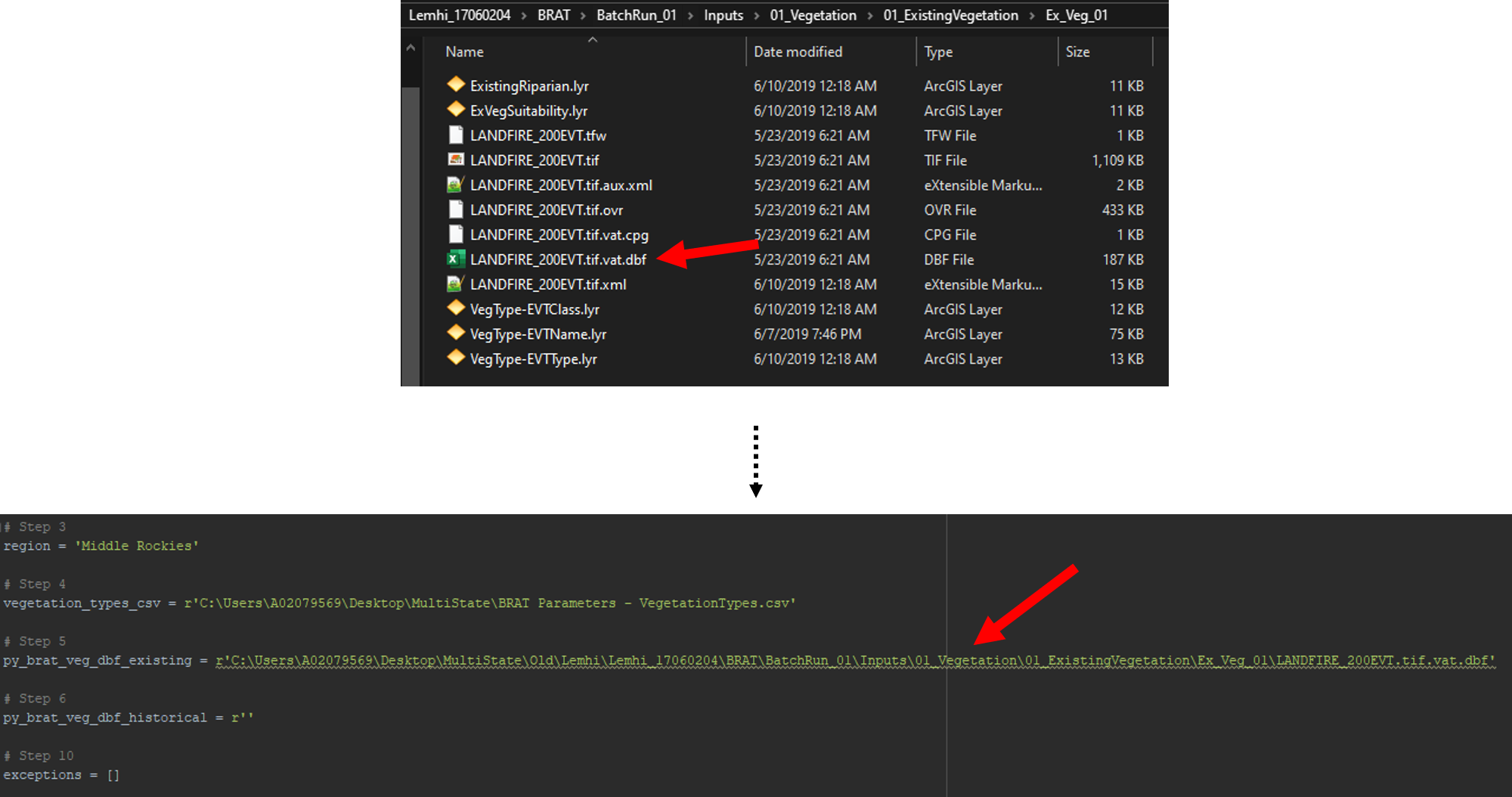
Step 6
Keep your pyBRAT project open to find the historic vegetation raster's .dbf file. This is typically found in Inputs -> 01_Vegetation -> 01_HistoricVegetation -> Hist_Veg_01. Find the Find the file that ends in .dbf. This will typically be something like LANDFIRE_200BPS.vat.dbf or LANDFIRE_140BPS.vat.dbf. Populate the script's fourth parameter (py_brat_veg_dbf_historical) with the path to that file.
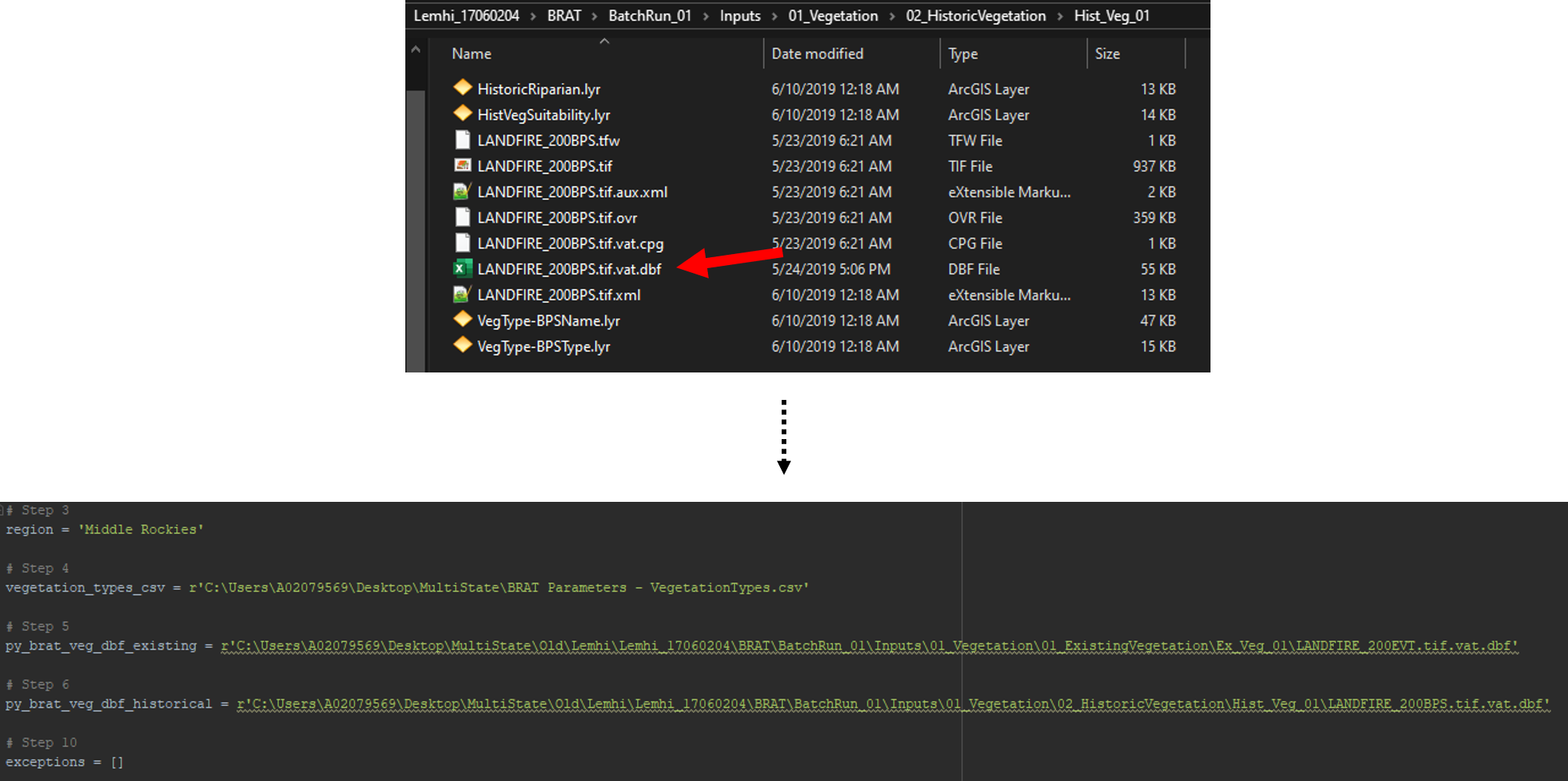
Step 7
Ensure that the script's fifth parameter (exceptions) is an empty list (Looks like []), then run the script. The script will print out several outputs into the console, so make sure you have that open. If your outputs are missing certain sections that's fine, as the script only outputs the sections if they have data you need to address.
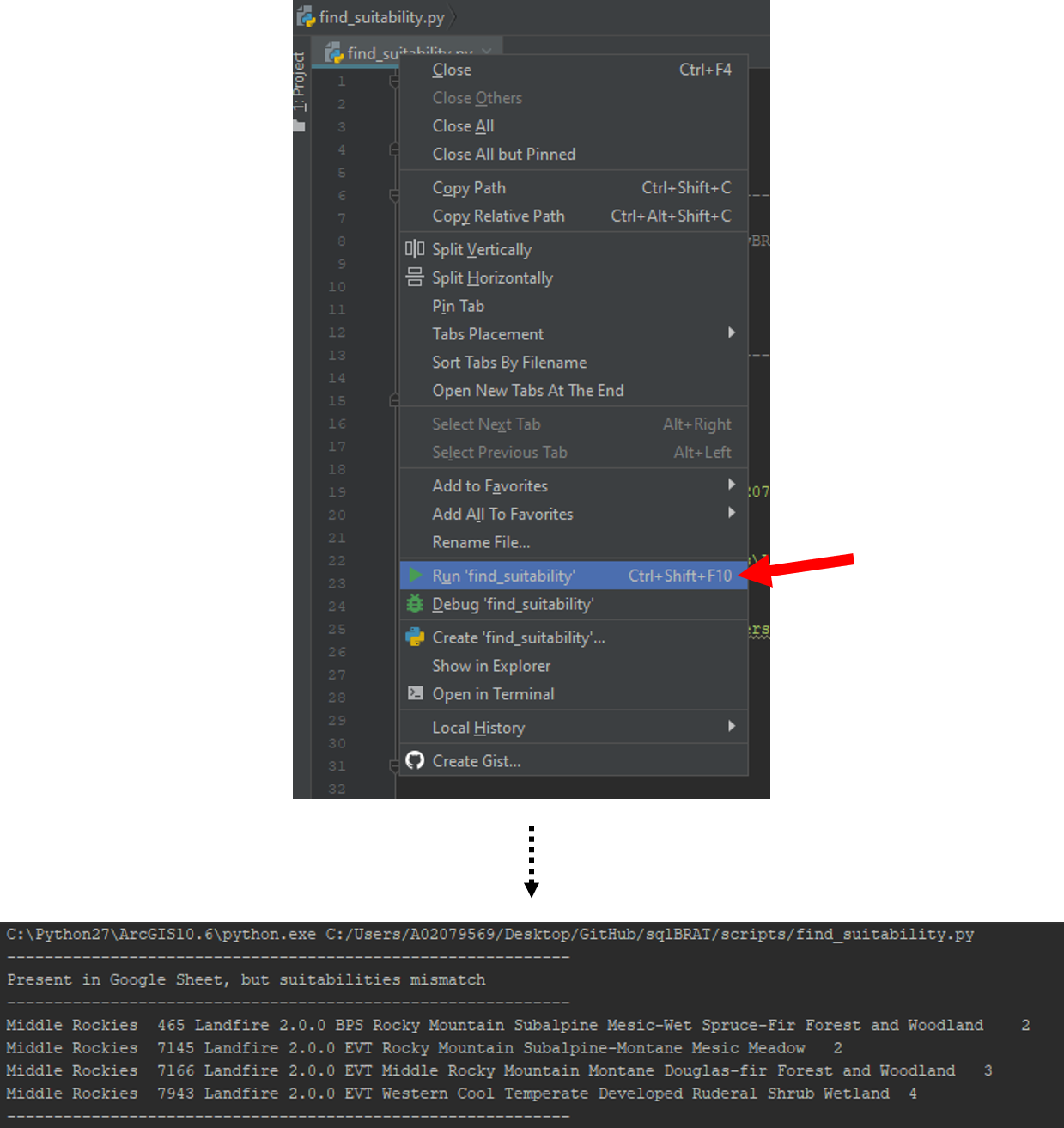
Step 8
The first section of the outputs we'll look at is titled "Present in Google Sheet, but default suitability should not be zero". All of the VegetationIDs listed here have default suitability values in the BRAT Parameters Google Sheet that need to be fixed. To do this, open the Google Sheet and navigate to the VegetationTypes tab. Find the vegetation ID that the first line of the script mentions, and replace its suitability with the one listed by the script. Repeat this for all values.
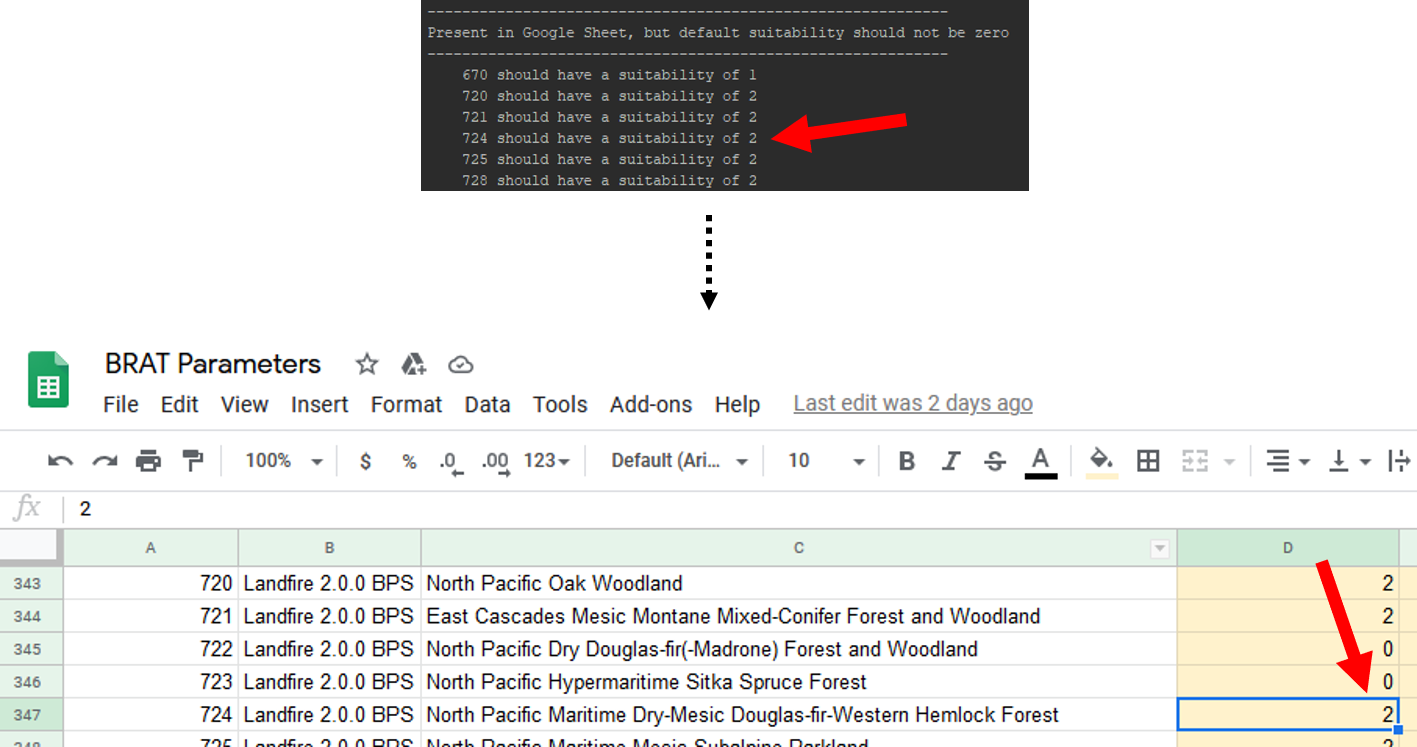
Step 9
The second section of the outputs we'll look at is titled "Present in Google Sheet, but suitabilities mismatch". All of the VegetationIDs listed here have default suitability values in the BRAT Parameters Google Sheet that don;t match the old pyBRAT values. To fix this, open the Google Sheet and navigate to the VegetationOverrides tab. Copy and paste all of the values from the script's output, into a blank section of the sheet. Ensure that there are no repeats (This can happen if someone has already entered suitability overrides for your region). Also ensure that there are no errors in the sheet, indicated by a red tag on the cell.
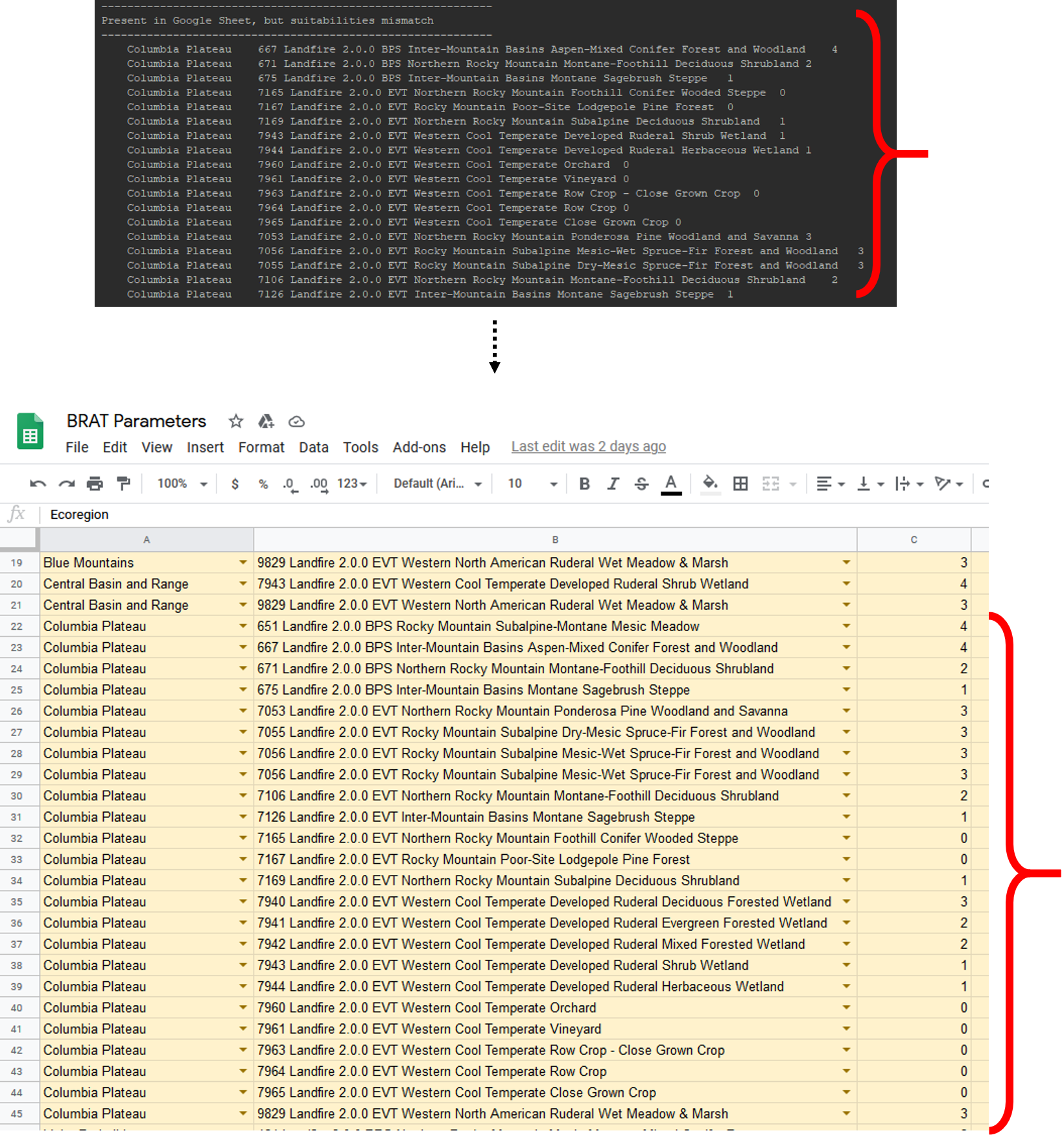
Step 10
The third section of the outputs we'll look at is titled "Present in Google Sheet, but names mismatch". This can happen if converting from LANDFIRE 1.4.0 values to LANDFIRE 2.0.0 values creates a mismatch in the name assigned to the same vegetationID. For each entry on this list, think critically to determine if the names truly represent different vegetation types.
If the two names truly represent different vegetation types:
This means there is an issue converting between LANDFIRE 1.4.0 values to LANDFIRE 2.0.0 values. If this happens, please report this, along with the VegetationID, LANDFIRE 1.4.0 name, and LANDFIRE 2.0.0 name, to this GitHub issue.
If the two names seem to represent the same vegetation type, and the difference between them is only semantic:
This means there is not really an issue converting between LANDFIRE 1.4.0 values to LANDFIRE 2.0.0 values. Add each vegetation ID that falls under this category to the exceptions parameter at the top of this script. Make sure entries are separated by a comma. After all exceptions have been added, rerun the script.
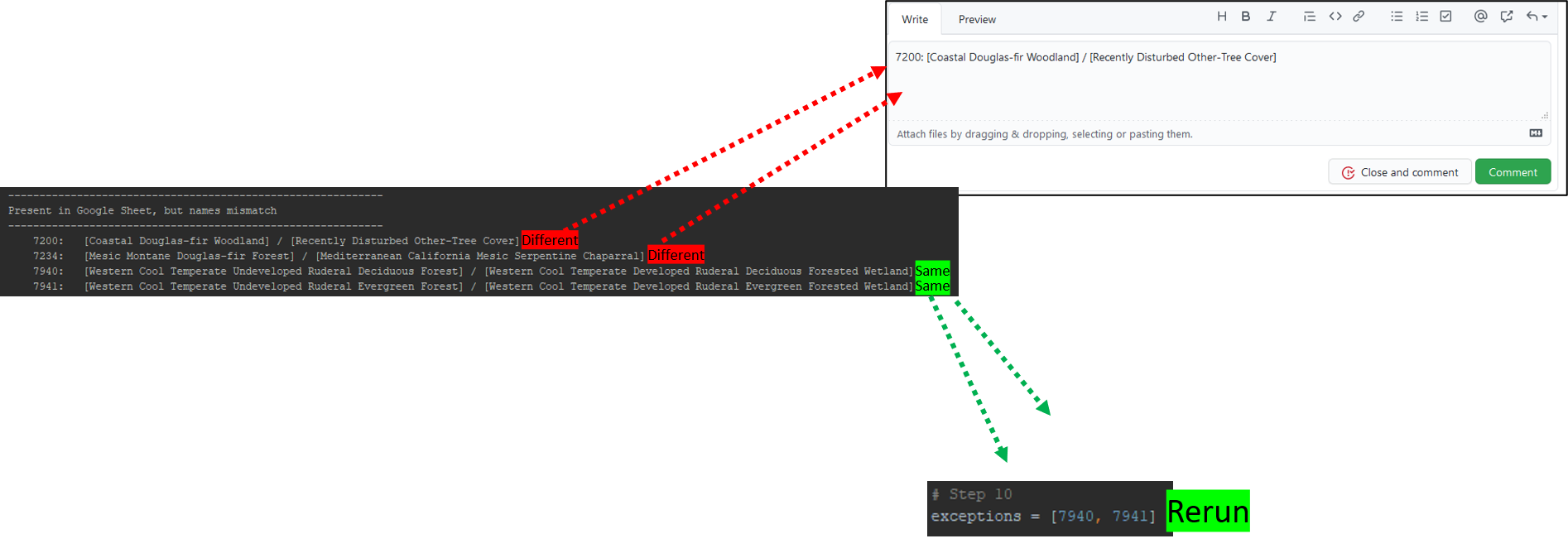
Step 11
The fourth and final section of the outputs we'll look at is titled "Not Present in Google Sheet". This means that a certain Vegetation ID is not present in Google Sheet, and thus has no default suitability. This can happen when LANDFIRE 1.4.0 values don't properly convert to 2.0.0. If this happens, please report this, along with the VegetationID, LANDFIRE name, epoch, and suitability value, to this GitHub issue.
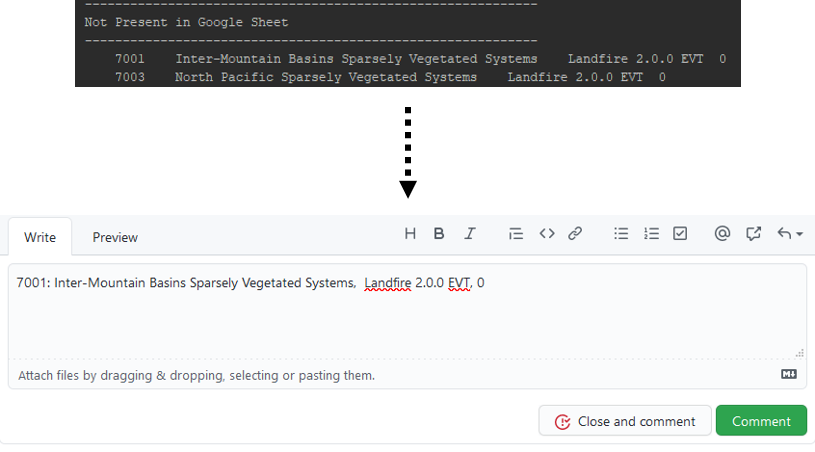
Step 12
That's everything! Now your sqlBRAT run will have appropriate values imported directly from and old pyBRAT project. It is still possible that there are some incorrect suitability values, so make sure that your rerun of sqlBRAT is still critically investigated for and suitability values that seem out of place.

